Unsupervised Defect Segmentation
Unsupervised Defect Segmentation can identify whether an object is in an abnormal state(NG), such as damage or deformation.
After completing the model annotations, refer to the video in the Training section to create dataset versions and train/deploy the model.
Use Case Scenarios
Unsupervised Defect Segmentation works on a single object. That is, the dataset should contain only one type of object, and the object's position must remain relatively fixed. The object will be classified as either normal or abnormal.
Unsupervised Defect Segmentation will learn to identify whether an object is in an abnormal state, using polygon annotations to mark the abnormal regions.
Annotation Method
If a pre-trained model exists, you can use the assisted annotation tool, allowing the deep learning model to help with annotations. You can then verify and correct the annotations as needed.
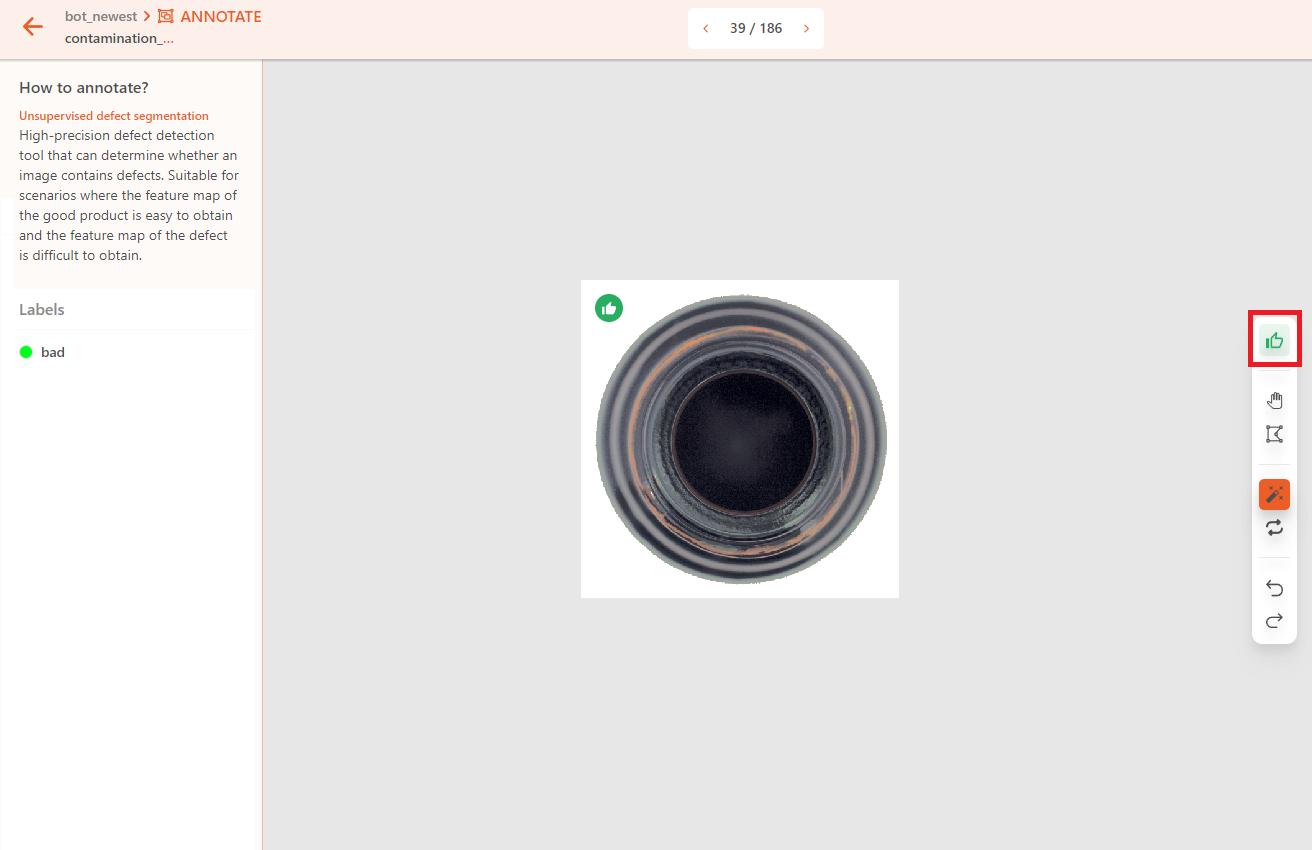
If the object has no defects, label it as good.
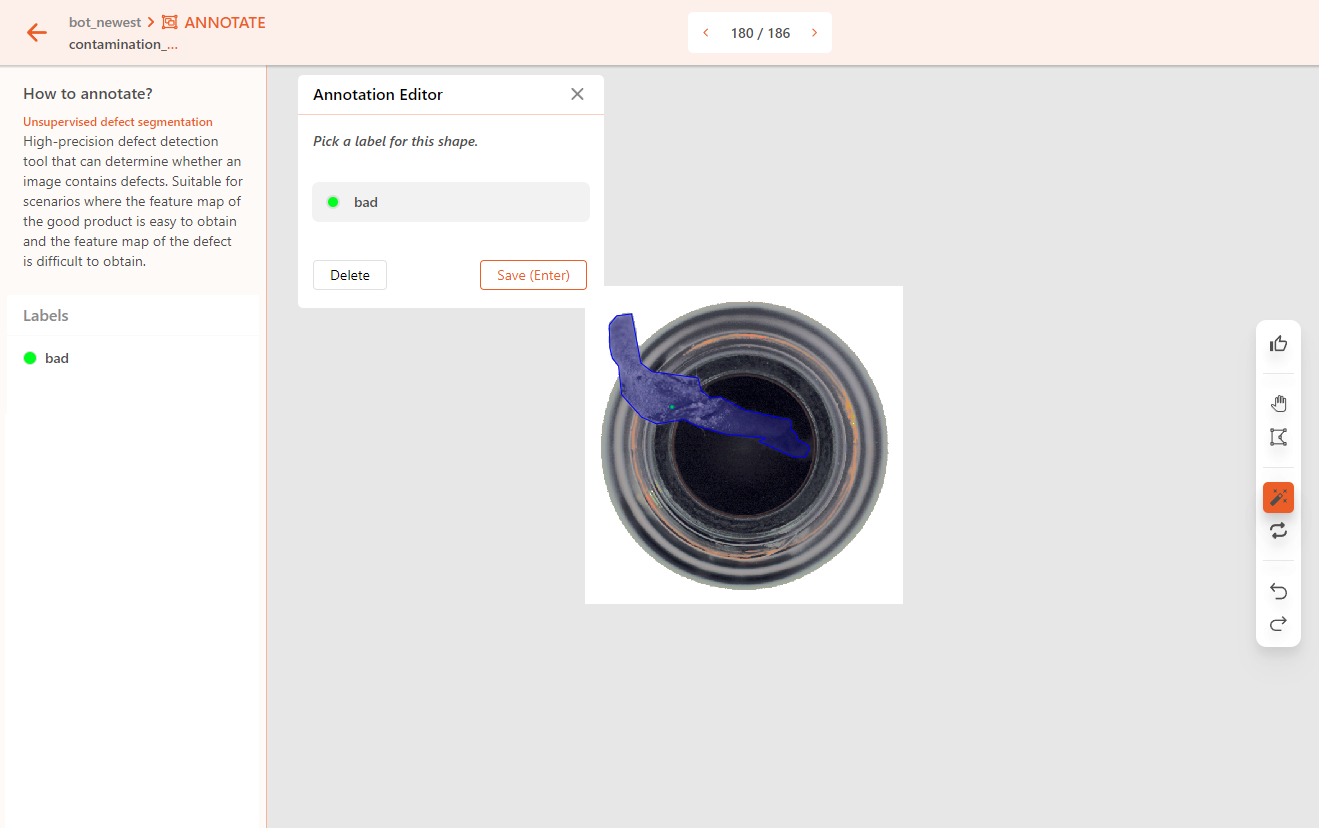
If the object has defects, mark it as abnormal. Use the polygon tool or smart polygon to annotate the outline of the defect area.
Notes
In an Unsupervised Defect Segmentation project, there should only be one label used to annotate the abnormal areas on an object.
Objects annotated as good should not include any defect region labels. Otherwise, it may lead to unsatisfactory training results or even training failure.
备注
During the training process of an Unsupervised Defect Segmentation project, ensure that the number of images assigned to the training set is less than or equal to the total number of defect-free images in the dataset. Otherwise, training may fail. Also, having too few defect images in the dataset may result in poor training outcomes.
Unlike other projects, Unsupervised Defect Segmentation does not apply any data augmentation options by default.
Practice
Download practice data with anomaly_detection.zip.
After unzipping, you will get 11 images and annotation (.json) files. Upload only the images to DaoAI World for annotation practice. Later, you can upload both images and annotation files to compare the results.